Page 35 - YORAM RUDY BOOK FINAL
P. 35
P. 3 5
P. 35
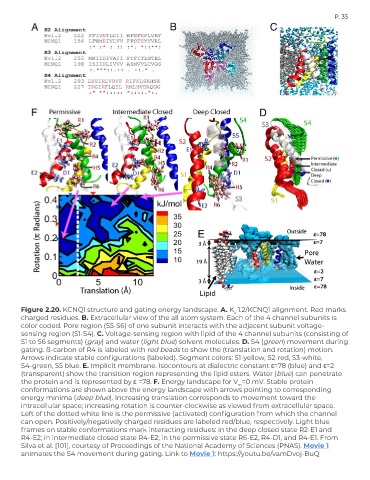
Figure 2.20. KCNQ1 structure and gating energy landscape. A. K 1.2/KCNQ1 alignment. Red marks
v
charged residues. B. Extracellular view of the all atom system. Each of the 4 channel subunits is
color coded. Pore region (S5-S6) of one subunit interacts with the adjacent subunit voltage-
sensing region (S1-S4). C. Voltage-sensing region with lipid of the 4 channel subunits (consisting of
S1 to S6 segments) (gray) and water (light blue) solvent molecules. D. S4 (green) movement during
gating. ß-carbon of R4 is labeled with red beads to show the (translation and rotation) motion.
Arrows indicate stable configurations (labeled). Segment colors: S1-yellow, S2-red, S3-white,
S4-green, S5 blue. E. Implicit membrane. Isocontours at dialectric constant ε=78 (blue) and ε=2
(transparent) show the transition region representing the lipid esters. Water (blue) can penetrate
the protein and is represented by ε =78. F. Energy landscape for V =0 mV. Stable protein
m
conformations are shown above the energy landscape with arrows pointing to corresponding
energy minima (deep blue). Increasing translation corresponds to movement toward the
intracellular space; increasing rotation is counter-clockwise as viewed from extracellular space.
Left of the dotted white line is the permissive (activated) configuration from which the channel
can open. Positively/negatively charged residues are labeled red/blue, respectively. Light blue
frames on stable conformations mark interacting residues: in the deep closed state R2-E1 and
R4-E2; in intermediate closed state R4-E2; in the permissive state R6-E2, R4-D1, and R4-E1. From
Silva et al. [101], courtesy of Proceedings of the National Academy of Sciences (PNAS). Movie 1
animates the S4 movement during gating. Link to Movie 1: https://youtu.be/vamDvoj-BuQ