Page 156 - Avian Virology: Current Research and Future Trends
P. 156
Infectious Bronchitis Virus | 149
works in the regulation of RNA synthesis remains to be deeply Assembly and release
investigated, host factors such as zinc finger CCHC-type and With the replication of the gRNA and the transcription of the
RNA-binding motif 1 (MADP1) is revealed to interact with sgRNAs, the viral proteins can now be translated using the host
the 5′ UTR of IBV RNA and enhance viral replication and translation machinery. Following translation, the S, E and M pro-
transcription (Tan et al., 2012). teins are co-translationally inserted into the ER. Moving along the
On the other hand, during RNA transcription, perhaps secretory pathway, these proteins will gather at the assembly point
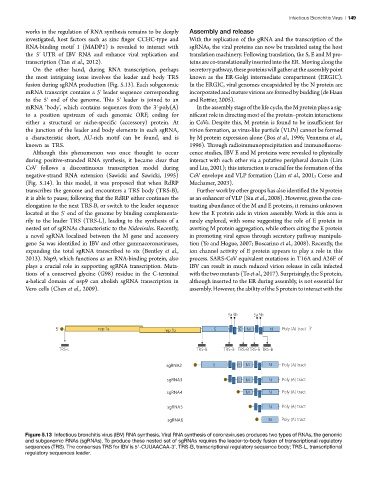
the most intriguing issue involves the leader and body TRS known as the ER-Golgi intermediate compartment (ERGIC).
fusion during sgRNA production (Fig. 5.13). Each subgenomic In the ERGIC, viral genomes encapsidated by the N protein are
mRNA transcript contains a 5′ leader sequence corresponding incorporated and mature virions are formed by budding (de Haan
to the 5′ end of the genome. This 5′ leader is joined to an and Rottier, 2005).
mRNA ‘body’, which contains sequences from the 3′-poly(A) In the assembly stage of the life cycle, the M protein plays a sig-
to a position upstream of each genomic ORF, coding for nificant role in directing most of the protein–protein interactions
either a structural or niche-specific (accessory) protein. At in CoVs. Despite this, M protein is found to be insufficient for
the junction of the leader and body elements in each sgRNA, virion formation, as virus-like particle (VLPs) cannot be formed
a characteristic short, AU-rich motif can be found, and is by M protein expression alone (Bos et al., 1996; Vennema et al.,
known as TRS. 1996). Through radioimmunoprecipitation and immunofluores-
Although this phenomenon was once thought to occur cence studies, IBV E and M proteins were revealed to physically
during positive-stranded RNA synthesis, it became clear that interact with each other via a putative peripheral domain (Lim
CoV follows a discontinuous transcription model during and Liu, 2001); this interaction is crucial for the formation of the
negative-strand RNA extension (Sawicki and Sawicki, 1995) CoV envelope and VLP formation (Lim et al., 2001; Corse and
(Fig. 5.14). In this model, it was proposed that when RdRP Machamer, 2003).
transcribes the genome and encounters a TRS body (TRS-B), Further work by other groups has also identified the N protein
it is able to pause; following that the RdRP either continues the as an enhancer of VLP (Siu et al., 2008). However, given the con-
elongation to the next TRS-B, or switch to the leader sequence trasting abundance of the M and E proteins, it remains unknown
located at the 5′ end of the genome by binding complementa- how the E protein aids in virion assembly. Work in this area is
rily to the leader TRS (TRS-L), leading to the synthesis of a rarely explored, with some suggesting the role of E protein in
nested set of sgRNAs characteristic to the Nidovirales. Recently, averting M protein aggregation, while others citing the E protein
a novel sgRNA localized between the M gene and accessory in promoting viral egress through secretory pathway manipula-
gene 5a was identified in IBV and other gammacoronaviruses, tion (Ye and Hogue, 2007; Boscarino et al., 2008). Recently, the
expanding the total sgRNA transcribed to six (Bentley et al., ion channel activity of E protein appears to play a role in this
2013). Nsp9, which functions as an RNA-binding protein, also process. SARS-CoV equivalent mutations in T16A and A26F of
plays a crucial role in supporting sgRNA transcription. Muta- IBV can result in much reduced virion release in cells infected
tions of a conserved glycine (G98) residue in the C-terminal with the two mutants (To et al., 2017). Surprisingly, the S protein,
α-helical domain of nsp9 can abolish sgRNA transcription in although inserted to the ER during assembly, is not essential for
Vero cells (Chen et al., 2009). assembly. However, the ability of the S protein to interact with the
Figure 5.13 Infectious bronchitis virus (IBV) RNA synthesis. Viral RNA synthesis of coronaviruses produces two types of RNAs, the genomic
Figure 13. Infectious bronchitis virus (IBV) RNA synthesis. Viral RNA synthesis of coronaviruses produces two types of RNAs, the genomic and subgenomic
and subgenomic RNAs (sgRNAs). To produce these nested set of sgRNAs requires the leader-to-body fusion of transcriptional regulatory
RNAs (sgRNAs). To produce these nested set of sgRNAs requires the leader-to-body fusion of transcriptional regulatory sequences (TRS). The consensus TRS
sequences (TRS). The consensus TRS for IBV is 5′-CUUAACAA-3’. TRS-B, transcriptional regulatory sequence body; TRS-L, transcriptional
for IBV is 5’-CUUAACAA-3’. TRS-L, transcriptional regulatory sequences leader; TRS-B, transcriptional regulatory sequence body.
regulatory sequences leader.