Page 308 - Linear Models for the Prediction of Animal Breeding Values 3rd Edition
P. 308
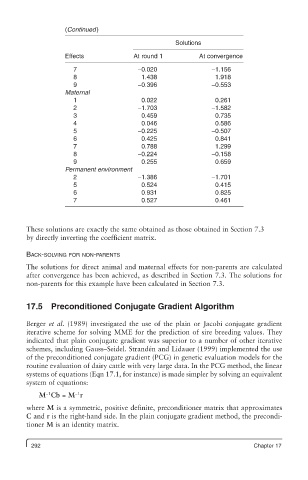
(Continued)
Solutions
Effects At round 1 At convergence
7 −0.020 −1.156
8 1.438 1.918
9 −0.396 −0.553
Maternal
1 0.022 0.261
2 −1.703 −1.582
3 0.459 0.735
4 0.046 0.586
5 −0.225 −0.507
6 0.425 0.841
7 0.788 1.299
8 −0.224 −0.158
9 0.255 0.659
Permanent environment
2 −1.386 −1.701
5 0.524 0.415
6 0.931 0.825
7 0.527 0.461
These solutions are exactly the same obtained as those obtained in Section 7.3
by directly inverting the coefficient matrix.
BACK-SOLVING FOR NON-PARENTS
The solutions for direct animal and maternal effects for non-parents are calculated
after convergence has been achieved, as described in Section 7.3. The solutions for
non-parents for this example have been calculated in Section 7.3.
17.5 Preconditioned Conjugate Gradient Algorithm
Berger et al. (1989) investigated the use of the plain or Jacobi conjugate gradient
iterative scheme for solving MME for the prediction of sire breeding values. They
indicated that plain conjugate gradient was superior to a number of other iterative
schemes, including Gauss–Seidel. Strandén and Lidauer (1999) implemented the use
of the preconditioned conjugate gradient (PCG) in genetic evaluation models for the
routine evaluation of dairy cattle with very large data. In the PCG method, the linear
systems of equations (Eqn 17.1, for instance) is made simpler by solving an equivalent
system of equations:
−1
−1
M Cb = M r
where M is a symmetric, positive definite, preconditioner matrix that approximates
C and r is the right-hand side. In the plain conjugate gradient method, the precondi-
tioner M is an identity matrix.
292 Chapter 17