Page 31 - Withrow and MacEwen's Small Animal Clinical Oncology, 6th Edition
P. 31
10 PART I The Biology and Pathogenesis of Cancer
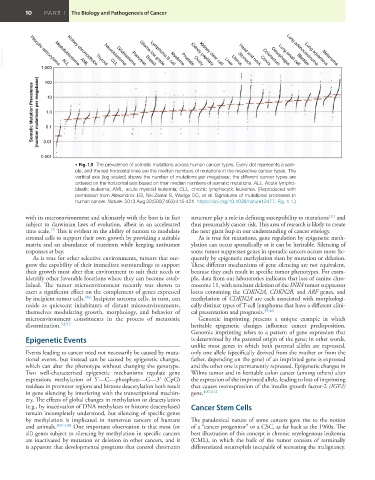
VetBooks.ir Pilocytic astrocytoma ALL AML Thyroid CLL Glioblastoma Glioma low grade Myeloma Kidney papillary Liver Uterus Cervix Oesophagus Bladder Melanoma
Neuroblastoma
Pancreas
Breast
Kidney chromophobe
Lung adenocarcinoma
Lung squamous
Medulloblastoma
Lymphoma B cell
Head and neck
Colorectum
Lung small cell
Stomach
Prostate
Ovary
Kidney clear cell
1,000
100
Somatic Mutation Prevalence (number mutations per megabase) 1.0
10
0.1
0.01
0.001
• Fig. 1.5 The prevalence of somatic mutations across human cancer types. Every dot represents a sam-
ple, and the red horizontal lines are the median numbers of mutations in the respective cancer types. The
vertical axis (log scaled) shows the number of mutations per megabase; the different cancer types are
ordered on the horizontal axis based on their median numbers of somatic mutations. ALL, Acute lympho-
blastic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia. (Reproduced with
permission from Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in
human cancer. Nature. 2013 Aug 22;500(7463):415-421. https://doi.org/10.1038/nature12477. Fig. 1.1.)
with its microenvironment and ultimately with the host is in fact structure play a role in defining susceptibility to mutations 111 and
subject to darwinian laws of evolution, albeit in an accelerated thus presumably cancer risk. This area of research is likely to create
time scale. This is evident in the ability of tumors to modulate the next giant leap in our understanding of cancer etiology.
75
stromal cells to support their own growth by providing a suitable As is true for mutations, gene regulation by epigenetic meth-
matrix and an abundance of nutrients while keeping antitumor ylation can occur sporadically or it can be heritable. Silencing of
responses at bay. some tumor suppressor genes in sporadic cancers occurs more fre-
As is true for other selective environments, tumors that out- quently by epigenetic methylation than by mutation or deletion.
grow the capability of their immediate surroundings to support These different mechanisms of gene silencing are not equivalent,
their growth must alter that environment to suit their needs or because they each result in specific tumor phenotypes. For exam-
identify other favorable locations where they can become estab- ple, data from our laboratories indicates that loss of canine chro-
lished. The tumor microenvironment recently was shown to mosome 11, with resultant deletion of the INK4 tumor suppressor
exert a significant effect on the complement of genes expressed locus containing the CDKN2A, CDKN2B, and ARF genes, and
by incipient tumor cells. 106 Incipient sarcoma cells, in turn, can methylation of CDKN2A are each associated with morphologi-
reside as quiescent inhabitants of distant microenvironments, cally distinct types of T-cell lymphoma that have a different clini-
themselves modulating growth, morphology, and behavior of cal presentation and prognosis. 39,40
microenvironment constituents in the process of metastatic Genomic imprinting presents a unique example in which
dissemination. 53,57 heritable epigenetic changes influence cancer predisposition.
Genomic imprinting refers to a pattern of gene expression that
Epigenetic Events is determined by the parental origin of the gene; in other words,
unlike most genes in which both parental alleles are expressed,
Events leading to cancer need not necessarily be caused by muta- only one allele (specifically derived from the mother or from the
tional events, but instead can be caused by epigenetic changes, father, depending on the gene) of an imprinted gene is expressed
which can alter the phenotype without changing the genotype. and the other one is permanently repressed. Epigenetic changes in
Two well-characterized epigenetic mechanisms regulate gene Wilms tumor and in heritable colon cancer (among others) alter
expression; methylation of 5’—C—phosphate—G—3’ (CpG) the expression of the imprinted allele, leading to loss of imprinting
residues in promoter regions and histone deacetylation both result that causes overexpression of the insulin growth factor-2 (IGF2)
in gene silencing by interfering with the transcriptional machin- gene. 107,112
ery. The effects of global changes in methylation or deacetylation
(e.g., by inactivation of DNA methylases or histone deacetylases) Cancer Stem Cells
remain incompletely understood, but silencing of specific genes
by methylation is implicated in numerous cancers of humans The paradoxical nature of some cancers gave rise to the notion
and animals. 107–110 One important observation is that most (or of a “cancer progenitor” or a CSC, as far back as the 1960s. The
all) genes subject to silencing by methylation in specific cancers best illustration of this concept is chronic myelogenous leukemia
are inactivated by mutation or deletion in other cancers, and it (CML), in which the bulk of the tumor consists of terminally
is apparent that developmental programs that control chromatin differentiated neutrophils incapable of recreating the malignancy.