Page 74 - Cardiac Electrophysiology | A Modeling and Imaging Approach
P. 74
P. 74
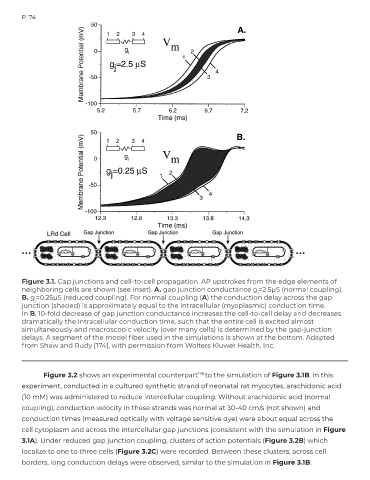
Figure 3.1. Gap junctions and cell-to-cell propagation. AP upstrokes from the edge elements of
neighboring cells are shown (see inset). A. gap junction conductance g =2.5μS (normal coupling).
j
B. g =0.25μS (reduced coupling). For normal coupling (A) the conduction delay across the gap
j
junction (shaded) is approximately equal to the intracellular (myoplasmic) conduction time.
In B, 10-fold decrease of gap junction conductance increases the cell-to-cell delay and decreases
dramatically the intracellular conduction time, such that the entire cell is excited almost
simultaneously and macroscopic velocity (over many cells) is determined by the gap-junction
delays. A segment of the model fiber used in the simulations is shown at the bottom. Adapted
from Shaw and Rudy [174], with permission from Wolters Kluwer Health, Inc.
Figure 3.2 shows an experimental counterpart 178 to the simulation of Figure 3.1B. In this
experiment, conducted in a cultured synthetic strand of neonatal rat myocytes, arachidonic acid
(10 mM) was administered to reduce intercellular coupling. Without arachidonic acid (normal
coupling), conduction velocity in these strands was normal at 30-40 cm/s (not shown) and
conduction times (measured optically with voltage sensitive dye) were about equal across the
cell cytoplasm and across the intercellular gap junctions (consistent with the simulation in Figure
3.1A). Under reduced gap junction coupling, clusters of action potentials (Figure 3.2B) which
localize to one to three cells (Figure 3.2C) were recorded. Between these clusters, across cell
borders, long conduction delays were observed, similar to the simulation in Figure 3.1B.