Page 47 - YORAM RUDY BOOK FINAL
P. 47
P. 47
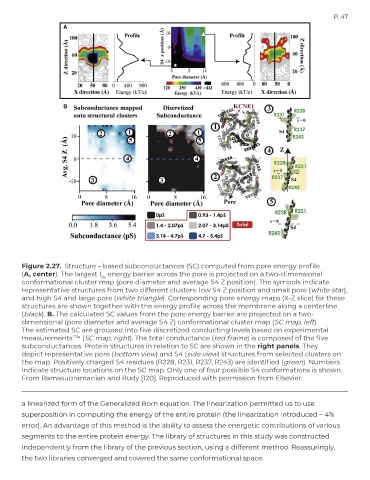
Figure 2.27. Structure – based subconductances (SC) computed from pore energy profile
(A, center). The largest I energy barrier across the pore is projected on a two-dimensional
Ks
conformational cluster map (pore diameter and average S4 Z position). The symbols indicate
representative structures from two different clusters: low S4 Z position and small pore (white star),
and high S4 and large pore (white triangle). Corresponding pore energy maps (X-Z slice) for these
structures are shown together with the energy profile across the membrane along a centerline
(black). B. The calculated SC values from the pore energy barrier are projected on a two-
dimensional (pore diameter and average S4 Z) conformational cluster map (SC map, left).
The estimated SC are grouped into five discretized conducting levels based on experimental
measurements 123a (SC map, right). The total conductance (red frame) is composed of the five
subconductances. Protein structures in relation to SC are shown in the right panels. They
depict representative pore (bottom view) and S4 (side view) structures from selected clusters on
the map. Positively charged S4 residues (R228, R231, R237, R243) are identified (green). Numbers
indicate structure locations on the SC map. Only one of four possible S4 conformations is shown.
From Ramasubramanian and Rudy [120]. Reproduced with permission from Elsevier.
a linearized form of the Generalized Born equation. The linearization permitted us to use
superposition in computing the energy of the entire protein (the linearization introduced ~ 4%
error). An advantage of this method is the ability to assess the energetic contributions of various
segments to the entire protein energy. The library of structures in this study was constructed
independently from the library of the previous section, using a different method. Reassuringly,
the two libraries converged and covered the same conformational space.