Page 62 - Withrow and MacEwen's Small Animal Clinical Oncology, 6th Edition
P. 62
CHAPTER 2 Tumor Biology and Metastasis 41
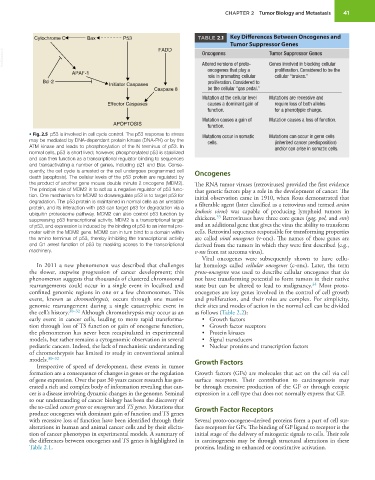
Cytochrome C Bax P53 TABLE 2.1 Key Differences Between Oncogenes and
Tumor Suppressor Genes
FADD
VetBooks.ir Oncogenes Tumor Suppressor Genes
Altered versions of proto-
Genes involved in blocking cellular
APAF-1 oncogenes that play a proliferation. Considered to be the
role in promoting cellular cellular “brakes.”
Bcl-2 proliferation. Considered to
Initiator Caspases
Caspase 8 be the cellular “gas pedal.”
Mutation at the cellular level Mutations are recessive and
Effector Caspases causes a dominant gain of require loss of both alleles
function. for a phenotypic change.
Mutation causes a gain of Mutation causes a loss of function.
APOPTOSIS function.
• Fig. 2.5 p53 is involved in cell cycle control. The p53 response to stress Mutations occur in somatic Mutations can occur in germ cells
may be mediated by DNA-dependent protein kinase (DNA-PK) or by the cells. (inherited cancer predisposition)
ATM kinase and leads to phosphorylation of the N terminus of p53. In and/or can arise in somatic cells.
normal cells, p53 is short lived; however, phosphorylated p53 is stabilized
and can then function as a transcriptional regulator binding to sequences
and transactivating a number of genes, including p21 and Bax. Conse-
quently, the cell cycle is arrested or the cell undergoes programmed cell Oncogenes
death (apoptosis). The cellular levels of the p53 protein are regulated by
the product of another gene mouse double minute 2 oncogene (MDM2). The RNA tumor viruses (retroviruses) provided the first evidence
The principal role of MDM2 is to act as a negative regulator of p53 func- that genetic factors play a role in the development of cancer. The
tion. One mechanism for MDM2 to downregulate p53 is to target p53 for initial observation came in 1910, when Rous demonstrated that
degradation. The p53 protein is maintained in normal cells as an unstable a filterable agent (later classified as a retrovirus and termed avian
protein, and its interaction with p53 can target p53 for degradation via a leukosis virus) was capable of producing lymphoid tumors in
ubiquitin proteosome pathway. MDM2 can also control p53 function by 33
suppressing p53 transcriptional activity. MDM2 is a transcriptional target chickens. Retroviruses have three core genes (gag, pol, and env)
of p53, and expression is induced by the binding of p53 to an internal pro- and an additional gene that gives the virus the ability to transform
moter within the MDM2 gene. MDM2 can in turn bind to a domain within cells. Retroviral sequences responsible for transforming properties
the amino terminus of p53, thereby inhibiting the transcriptional activity are called viral oncogenes (v-onc). The names of these genes are
and G1 arrest function of p53 by masking access to the transcriptional derived from the tumors in which they were first described (e.g.,
machinery. v-ras from rat sarcoma virus).
Viral oncogenes were subsequently shown to have cellu-
In 2011 a new phenomenon was described that challenges lar homologs called cellular oncogenes (c-onc). Later, the term
the slower, stepwise progression of cancer development; this proto-oncogene was used to describe cellular oncogenes that do
phenomenon suggests that thousands of clustered chromosomal not have transforming potential to form tumors in their native
rearrangements could occur in a single event in localized and state but can be altered to lead to malignancy. Most proto-
34
confined genomic regions in one or a few chromosomes. This oncogenes are key genes involved in the control of cell growth
event, known as chromothrypsis, occurs through one massive and proliferation, and their roles are complex. For simplicity,
genomic rearrangement during a single catastrophic event in their sites and modes of action in the normal cell can be divided
the cell’s history. 30–32 Although chromothrypsis may occur as an as follows (Table 2.2):
early event in cancer cells, leading to more rapid transforma- • Growth factors
tion through loss of TS function or gain of oncogene function, • Growth factor receptors
the phenomenon has never been recapitulated in experimental • Protein kinases
models, but rather remains a cytogenomic observation in several • Signal transducers
pediatric cancers. Indeed, the lack of mechanistic understanding • Nuclear proteins and transcription factors
of chromothrypsis has limited its study in conventional animal
models. 30–32 Growth Factors
Irrespective of speed of development, these events in tumor
formation are a consequence of changes in genes or the regulation Growth factors (GFs) are molecules that act on the cell via cell
of gene expression. Over the past 30 years cancer research has gen- surface receptors. Their contribution to carcinogenesis may
erated a rich and complex body of information revealing that can- be through excessive production of the GF or through ectopic
cer is a disease involving dynamic changes in the genome. Seminal expression in a cell type that does not normally express that GF.
to our understanding of cancer biology has been the discovery of
the so-called cancer genes or oncogenes and TS genes. Mutations that Growth Factor Receptors
produce oncogenes with dominant gain of function and TS genes
with recessive loss of function have been identified through their Several proto-oncogene–derived proteins form a part of cell sur-
alterations in human and animal cancer cells and by their elicita- face receptors for GFs. The binding of GF ligand to receptor is the
tion of cancer phenotypes in experimental models. A summary of initial stage of the delivery of mitogenic signals to cells. Their role
the differences between oncogenes and TS genes is highlighted in in carcinogenesis may be through structural alterations in these
Table 2.1. proteins, leading to enhanced or constitutive activation.