Page 53 - Тахианаас хүнсний хордлого үүсгэгч CAMPYLOBACTER SPP.-ийг илрүүлсэн судалгааны дүн
P. 53
[ШИНЖЛЭХ УХААНЫ МАГИСТРЫН ЗЭРЭГ ГОРИЛСОН БҮТЭЭЛ] Хураангуй
Дүгнэлт
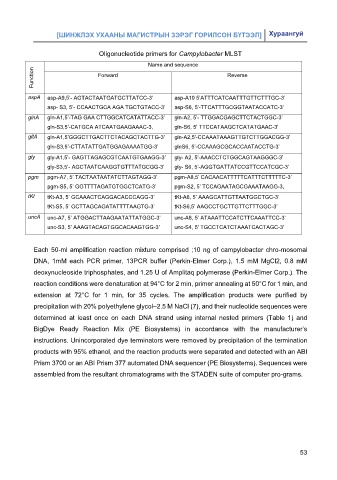
Oligonucleotide primers for Campylobacter MLST
Name and sequence
Function Forward Reverse
aspA asp-A9,5’- AGTACTAATGATGCTTATCC-3’ asp-A10 5’ATTTCATCAATTTGTTCTTTGC-3’
asp- S3, 5’- CCAACTGCA AGA TGCTGTACC-3’ asp-S6, 5’-TTCATTTGCGGTAATACCATC-3’
glnA gln-A1,5’-TAG GAA CTTGGCATCATATTACC-3’ gln-A2, 5’- TTGGACGAGCTTCTACTGGC-3’
gln-S3,5’-CATGCA ATCAATGAAGAAAC-3, gln-S6, 5’ TTCCATAAGCTCATATGAAC-3’
gltA gln-A1,5’GGGCTTGACTTCTACAGCTACTTG-3’ gln-A2,5’-CCAAATAAAGTTGTCTTGGACGG-3’
gln-S3,5’-CTTATATTGATGGAGAAAATGG-3’ glnS6, 5’-CCAAAGCGCACCAATACCTG-3’
gly gly-A1,5’- GAGTTAGAGCGTCAATGTGAAGG-3’ gly- A2, 5’-AAACCTCTGGCAGTAAGGGC-3’
gly-S3,5’- AGCTAATCAAGGTGTTTATGCGG-3’ gly- S6, 5’-AGGTGATTATCCGTTCCATCGC-3’
pgm pgm-A7, 5’ TACTAATAATATCTTAGTAGG-3’ pgm-A8,5’ CACAACATTTTTCATTTCTTTTTC-3’
pgm-S5, 5’ GGTTTTAGATGTGGCTCATG-3’ pgm-S2, 5’ TCCAGAATAGCGAAATAAGG-3,
tKt tKt-A3, 5’ GCAAACTCAGGACACCCAGG-3’ tKt-A6, 5’ AAAGCATTGTTAATGGCTGC-3’
tKt-S5, 5’ GCTTAGCAGATATTTTAAGTG-3’ tKt-S6,5’ AAGCCTGCTTGTTCTTTGGC-3’
uncA unc-A7, 5’ ATGGACTTAAGAATATTATGGC-3’ unc-A8, 5’ ATAAATTCCATCTTCAAATTCC-3’
unc-S3, 5’ AAAGTACAGTGGCACAAGTGG-3’ unc-S4, 5’ TGCCTCATCTAAATCACTAGC-3’
Each 50-ml amplification reaction mixture comprised ;10 ng of campylobacter chro-mosomal
DNA, 1mM each PCR primer, 13PCR buffer (Perkin-Elmer Corp.), 1.5 mM MgCl2, 0.8 mM
deoxynucleoside triphosphates, and 1.25 U of Amplitaq polymerase (Perkin-Elmer Corp.). The
reaction conditions were denaturation at 94°C for 2 min, primer annealing at 50°C for 1 min, and
extension at 72°C for 1 min, for 35 cycles. The amplification products were purified by
precipitation with 20% polyethylene glycol–2.5 M NaCl (7), and their nucleotide sequences were
determined at least once on each DNA strand using internal nested primers (Table 1) and
BigDye Ready Reaction Mix (PE Biosystems) in accordance with the manufacturer’s
instructions. Unincorporated dye terminators were removed by precipitation of the termination
products with 95% ethanol, and the reaction products were separated and detected with an ABI
Prism 3700 or an ABI Prism 377 automated DNA sequencer (PE Biosystems). Sequences were
assembled from the resultant chromatograms with the STADEN suite of computer pro-grams.
53