Page 57 - Avian Virology: Current Research and Future Trends
P. 57
50 | Samal
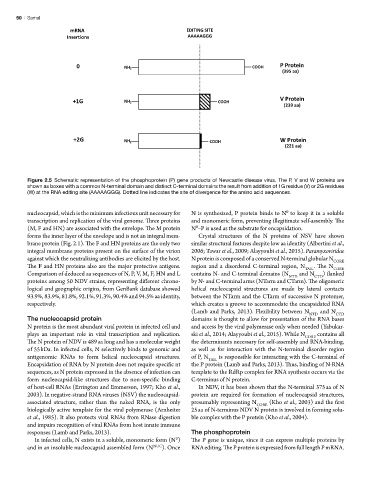
mRNA EDITING SITE
Insertions AAAAAGGG
0 NH 2 COOH P Protein
(395 aa)
+1G NH 2 COOH V Protein
(239 aa)
+2G NH 2 COOH W Protein
(221 aa)
Figure 2.5 Schematic representation of the phosphoprotein (P) gene products of Newcastle disease virus. The P, V and W proteins are
shown as boxes with a common N-terminal domain and distinct C-terminal domains the result from addition of 1G residue (V) or 2G residues
Figure 5
(W) at the RNA editing site (AAAAAGGG). Dotted line indicates the site of divergence for the amino acid sequences.
nucleocapsid, which is the minimum infectious unit necessary for N is synthesized, P protein binds to N to keep it in a soluble
0
transcription and replication of the viral genome. Three proteins and monomeric form, preventing illegitimate self-assembly. The
(M, F and HN) are associated with the envelope. The M protein N –P is used as the substrate for encapsidation.
0
forms the inner layer of the envelope and is not an integral mem- Crystal structures of the N proteins of NSV have shown
brane protein (Fig. 2.1). The F and HN proteins are the only two similar structural features despite low aa identity (Albertini et al.,
integral membrane proteins present on the surface of the virion 2006; Tawar et al., 2009; Alayyoubi et al., 2015). Paramyxoviridae
against which the neutralizing antibodies are elicited by the host. N protein is composed of a conserved N-terminal globular N CORE
The F and HN proteins also are the major protective antigens. region and a disordered C-terminal region, N TAIL . The N CORE
Comparison of deduced aa sequences of N, P, V, M, F, HN and L contains N- and C-terminal domains (N NTD and N CTD ) flanked
proteins among 50 NDV strains, representing different chrono- by N- and C-terminal arms (NTarm and CTarm). The oligomeric
logical and geographic origins, from GenBank database showed helical nucleocapsid structures are made by lateral contacts
93.9%, 83.9%, 81.8%, 92.1%, 91.3%, 90.4% and 94.5% aa identity, between the NTarm and the CTarm of successive N protomer,
respectively. which creates a groove to accommodate the encapsidated RNA
(Lamb and Parks, 2013). Flexibility between N NTD and N CTD
The nucleocapsid protein domains is thought to allow for presentation of the RNA bases
N protein is the most abundant viral protein in infected cell and and access by the viral polymerase only when needed (Yabukar-
plays an important role in viral transcription and replication. ski et al., 2014; Alayyoubi et al., 2015). While N CORE contains all
The N protein of NDV is 489 aa long and has a molecular weight the determinants necessary for self-assembly and RNA-binding,
of 55 kDa. In infected cells, N selectively binds to genomic and as well as for interaction with the N-terminal disorder region
antigenomic RNAs to form helical nucleocapsid structures. of P, N TAIL is responsible for interacting with the C-terminal of
Encapsidation of RNA by N protein does not require specific nt the P protein (Lamb and Parks, 2013). Thus, binding of N-RNA
sequences, as N protein expressed in the absence of infection can template to the RdRp complex for RNA synthesis occurs via the
form nucleocapsid-like structures due to non-specific binding C-terminus of N protein.
of host-cell RNAs (Errington and Emmerson, 1997; Kho et al., In NDV, it has been shown that the N-terminal 375 aa of N
2003). In negative-strand RNA viruses (NSV) the nucleocapsid- protein are required for formation of nucleocapsid structures,
associated structure, rather than the naked RNA, is the only presumably representing N CORE (Kho et al., 2003) and the first
biologically active template for the viral polymerase (Arnheiter 25 aa of N-terminus NDV N protein is involved in forming solu-
et al., 1985). It also protects viral RNAs from RNase digestion ble complex with the P protein (Kho et al., 2004).
and impairs recognition of viral RNAs from host innate immune
responses (Lamb and Parks, 2013). The phosphoprotein
In infected cells, N exists in a soluble, monomeric form (N ) The P gene is unique, since it can express multiple proteins by
0
and in an insoluble nucleocapsid assembled form (N NUC ). Once RNA editing. The P protein is expressed from full length P mRNA.