Page 56 - YORAM RUDY BOOK FINAL
P. 56
P. 56
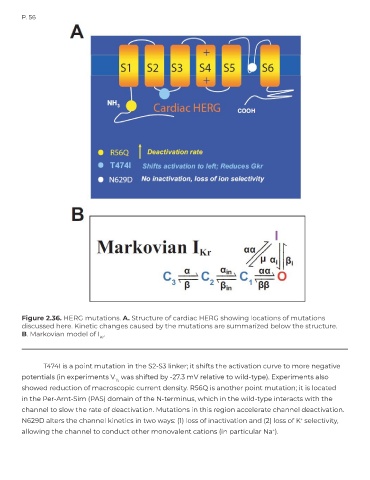
Figure 2.36. HERG mutations. A. Structure of cardiac HERG showing locations of mutations
discussed here. Kinetic changes caused by the mutations are summarized below the structure.
B. Markovian model of I .
Kr
T474I is a point mutation in the S2-S3 linker; it shifts the activation curve to more negative
potentials (in experiments V was shifted by -27.3 mV relative to wild-type). Experiments also
½
showed reduction of macroscopic current density. R56Q is another point mutation; it is located
in the Per-Arnt-Sim (PAS) domain of the N-terminus, which in the wild-type interacts with the
channel to slow the rate of deactivation. Mutations in this region accelerate channel deactivation.
N629D alters the channel kinetics in two ways: (1) loss of inactivation and (2) loss of K selectivity,
+
allowing the channel to conduct other monovalent cations (in particular Na ).
+