Page 566 - The Toxicology of Fishes
P. 566
546 The Toxicology of Fishes
C
O H 3
H 3 C
Enzymatic O
N N N N
Hydroxylation
C
H 3 H CH C
3
2
OH
CH O + H O
2
2
CH N +
3 2
+
OCH 3
O
HN
N HN
N
NH 2 N
N NH 2 N
N
6
O -Methylguanine Guanine
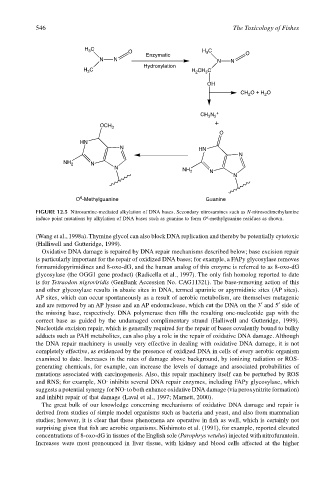
FIGURE 12.5 Nitrosamine-mediated alkylation of DNA bases. Secondary nitrosamines such as N-nitrosodimethylamine
6
induce point mutations by alkylation of DNA bases such as guanine to form O -methylguanine residues as shown.
(Wang et al., 1998a). Thymine glycol can also block DNA replication and thereby be potentially cytotoxic
(Halliwell and Gutteridge, 1999).
Oxidative DNA damage is repaired by DNA repair mechanisms described below; base excision repair
is particularly important for the repair of oxidized DNA bases; for example, a FAPy glycosylase removes
formamidopyrimidines and 8-oxo-dG, and the human analog of this enzyme is referred to as 8-oxo-dG
glycosylase (the OGG1 gene product) (Radicella et al., 1997). The only fish homolog reported to date
is for Tetraodon nigroviridis (GenBank Accession No. CAG11321). The base-removing action of this
and other glycosylase results in abasic sites in DNA, termed apurinic or apyrmidinic sites (AP sites).
AP sites, which can occur spontaneously as a result of aerobic metabolism, are themselves mutagenic
and are removed by an AP lysase and an AP endonuclease, which cut the DNA on the 3′ and 5′ side of
the missing base, respectively. DNA polymerase then fills the resulting one-nucleotide gap with the
correct base as guided by the undamaged complimentary strand (Halliwell and Gutteridge, 1999).
Nucleotide excision repair, which is generally required for the repair of bases covalently bound to bulky
adducts such as PAH metabolites, can also play a role in the repair of oxidative DNA damage. Although
the DNA repair machinery is usually very effective in dealing with oxidative DNA damage, it is not
completely effective, as evidenced by the presence of oxidized DNA in cells of every aerobic organism
examined to date. Increases in the rates of damage above background, by ionizing radiation or ROS-
generating chemicals, for example, can increase the levels of damage and associated probabilities of
mutations associated with carcinogenesis. Also, this repair machinery itself can be perturbed by ROS
and RNS; for example, NO· inhibits several DNA repair enzymes, including FAPy glycosylase, which
suggests a potential synergy for NO· to both enhance oxidative DNA damage (via peroxynitrite formation)
and inhibit repair of that damage (Laval et al., 1997; Marnett, 2000).
The great bulk of our knowledge concerning mechanisms of oxidative DNA damage and repair is
derived from studies of simple model organisms such as bacteria and yeast, and also from mammalian
studies; however, it is clear that these phenomena are operative in fish as well, which is certainly not
surprising given that fish are aerobic organisms. Nishimoto et al. (1991), for example, reported elevated
concentrations of 8-oxo-dG in tissues of the English sole (Parophrys vetulus) injected with nitrofurantoin.
Increases were most pronounced in liver tissue, with kidney and blood cells affected at the higher