Page 381 - Veterinary Toxicology, Basic and Clinical Principles, 3rd Edition
P. 381
348 SECTION | III Nanoparticles, Radiation and Carcinogens
VetBooks.ir protein complex, (2) unwinding of the DNA duplex
around the damage, (3) dual incision of the damaged
DNA strand to remove 30 or more nucleotides creating a
gap, (4) gap repair synthesis by DNA polymerase, and (5)
sealing of the gaps by ligase. The damage recognition in
transcriptionally silent region versus transcriptionally
active region requires different sets of proteins, but the
subsequent steps are essentially identical (Fig. 20.5D).
The MMR mechanism repairs bases that violate
Watson-Crick base pairing rules. The classic example is
that of Escherichia coli. The sequence 5 -GATC-3 in
0
0
E. coli DNA is methylated at adenine, and the sequences
0
0
0
5 -CCAGG-3 and 5 -CCTGG-3 are methylated at cyto-
0
sine. When DNA replicates, the daughter strand methyla-
tion is delayed. As a result, the newly synthesized
daughter strand remains undermethylated for some time
compared to the parental strand. If there is a base misin-
corporation, the MMR machinery (MutS-MutL-MutH
protein complex) identifies the misincorporated base by
scanning the methylation status of both strands. The
mismatched base is excised from the undermethylated
daughter strand.
DNA strand breaks are frequently caused by ionizing
radiation and chemicals that generate free radicals. Single
strand breaks do not disrupt the integrity of the DNA. The
intact single strand is coated by Poly(ADP-ribose)
polymerase-1 (PARP 1) protein near the lesion site of the
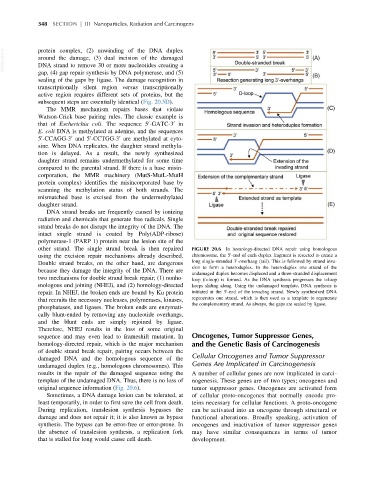
other strand. The single strand break is then repaired FIGURE 20.6 In homology-directed DNA repair using homologous
0
using the excision repair mechanisms already described. chromosome, the 5 -end of each duplex fragment is resected to create a
0
Double strand breaks, on the other hand, are dangerous long single-stranded 3 -overhang (tail). This is followed by strand inva-
sion to form a heteroduplex. In the heteroduplex one strand of the
because they damage the integrity of the DNA. There are
undamaged duplex becomes displaced and a three-stranded displacement
two mechanisms for double strand break repair; (1) nonho- loop (D-loop) is formed. As the DNA synthesis progresses the D-loop
mologous end joining (NHEJ), and (2) homology-directed keeps sliding along. Using the undamaged template, DNA synthesis is
0
repair. In NHEJ, the broken ends are bound by Ku protein initiated at the 3 -end of the invading strand. Newly synthesized DNA
that recruits the necessary nucleases, polymerases, kinases, regenerates one strand, which is then used as a template to regenerate
the complementary strand. As always, the gaps are sealed by ligase.
phosphatases, and ligases. The broken ends are enzymati-
cally blunt-ended by removing any nucleotide overhangs,
and the blunt ends are simply rejoined by ligase.
Therefore, NHEJ results in the loss of some original
sequence and may even lead to frameshift mutation. In Oncogenes, Tumor Suppressor Genes,
homology-directed repair, which is the major mechanism and the Genetic Basis of Carcinogenesis
of double strand break repair, pairing occurs between the
damaged DNA and the homologous sequence of the Cellular Oncogenes and Tumor Suppressor
undamaged duplex (e.g., homologous chromosomes). This Genes Are Implicated in Carcinogenesis
results in the repair of the damaged sequence using the A number of cellular genes are now implicated in carci-
template of the undamaged DNA. Thus, there is no loss of nogenesis. These genes are of two types; oncogenes and
original sequence information (Fig. 20.6). tumor suppressor genes. Oncogenes are activated form
Sometimes, a DNA damage lesion can be tolerated, at of cellular proto-oncogenes that normally encode pro-
least temporarily, in order to first save the cell from death. teins necessary for cellular functions. A proto-oncogene
During replication, translesion synthesis bypasses the can be activated into an oncogene through structural or
damage and does not repair it; it is also known as bypass functional alterations. Broadly speaking, activation of
synthesis. The bypass can be error-free or error-prone. In oncogenes and inactivation of tumor suppressor genes
the absence of translesion synthesis, a replication fork may have similar consequences in terms of tumor
that is stalled for long would cause cell death. development.