Page 118 - Avian Virology: Current Research and Future Trends
P. 118
Avian Paramyxoviruses | 111
the Western Hemisphere and the Eastern Hemisphere lineages Reverse genetics
(Choi et al., 2013; Wang et al., 2013). Based on F gene sequence, The method of understanding the functions of a gene by analys-
APMV-4 strains were classified into five clades (A to E) that were ing the phenotypic effects of specific mutations is called reverse
almost exclusively monophyletic with respect to the continent of genetics. To make reverse genetic studies possible for a RNA
origin (Reeves et al., 2016). Based on mean inter-populational virus, a system must be first developed to produce an infectious
evolutionary distances of the full F protein at least three geno- virus from cloned cDNA. The method to produce an infectious
types (I, II, and III) were found to exist within APMV-4 (Yin et negative-sense RNA virus entirely from cloned cDNA was first
al., 2017). Genotype I contained isolates mainly from old world developed for the Rabies virus (Schnell et al., 1994). Since
countries (Europe, Asia, and Africa), whereas genotypes II and then the same experimental approach has been used to recover
III comprise viruses originating from waterfowl from the USA other viruses of the order Mononegavirales from cloned cDNAs.
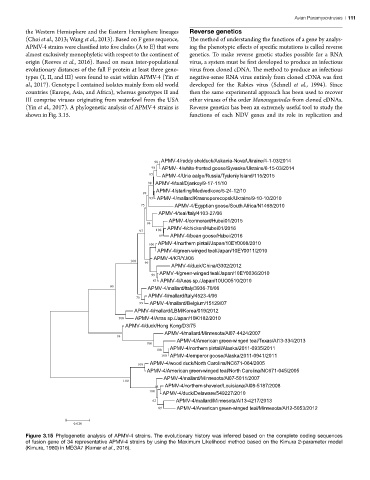
(Yin et al., 2017). A phylogenetic analysis of APMV-4 strains is Reverse genetics has been an extremely useful tool to study the
shown in Fig. 3.15. functions of each NDV genes and its role in replication and
Figure 15 98 APMV-4/ruddy shelduck/Askania-Nova/Ukraine/4-1-03/2014
98 APMV- 4/white-fronted goose/Syvaske/Ukraine/6-15-03/2014
65 APMV-4/Uria aalge/Russia/Tyuleniy Island/115/2015
98 APMV-4/teal/Djankoy/9-17-11/10
APMV-4/starling/Medvedkovo/5-24-12/10
89
93 APMV-4/mallard/Krasnoperecopsk/Ukraine/9-10-10/2010
75 APMV-4/Egyptian goose/South Africa/N1468/2010
APMV-4/teal/Italy/4103-27/06
APMV-4/cormorant/Hubei01/2015
99
97 100 APMV-4/chicken/Hubei01/2016
69 APMV-4/bean goose/Hubei/2016
100 APMV-4/northern pintail/Japan/10EY0008/2010
APMV-4/green-winged teal/Japan/10EY0011/2010
APMV-4/KR/YJ/06
100 99
APMV-4/duck/China/G302/2012
APMV-4/green-winged teal/Japan/10EY0036/2010
99
83 APMV-4/Anas sp./Japan/10UO0510/2010
99 APMV-4/mallard/Italy/3936-70/06
APMV-4/mallard/Italy/4523-4/06
76
99 APMV-4/mallard/Belgium/15129/07
APMV-4/mallard/LBM/Korea/019/2012
100 APMV-4/Anas sp./Japan/10KI182/2010
APMV-4/duck/Hong Kong/D3/75
APMV-4/mallard/Minnesota/AI07-4424/2007
99
APMV-4/American green-winged teal/Texas/AI13-334/2013
100
100 APMV-4/northern pintail/Alaska/2011-0935/2011
100 APMV-4/emperor goose/Alaska/2011-0941/2011
100 APMV-4/wood duck/North Carolina/NC671-064/2005
APMV-4/American green-winged teal/North Carolina/NC671-045/2005
APMV-4/mallard/Minnesota/AI07-5011/2007
100
93 APMV-4/northern shoveler/Louisiana/AI08-5187/2008
100 APMV-4/duck/Delaware/549227/2010
62 APMV-4/mallard/Minnesota/AI13-4217/2013
89 APMV-4/American green-winged teal/Minnesota/AI12-5053/2012
0.020
Figure 3.15 Phylogenetic analysis of APMV-4 strains. The evolutionary history was inferred based on the complete coding sequences
of fusion gene of 34 representative APMV-4 strains by using the Maximum Likelihood method based on the Kimura 2-parameter model
(Kimura, 1980) in MEGA7 (Kumar et al., 2016).