Page 173 - Veterinary Toxicology, Basic and Clinical Principles, 3rd Edition
P. 173
140 SECTION | I General
VetBooks.ir Parameter Estimation and Identifiability parameters. To reduce the likelihood of this type of error,
multiple tissue compartments may be sampled. Thus, it is
Accurate parameter values are essential for PBPK models
not only the number of data points representing the
to achieve their full predictive potential. The relevant para-
final model output that are used, but also the number of
meters are often physiological (blood flow, organ volume,
sampled compartments that is important for accurate
vascular space volume, etc.), and physicochemical (parti-
parameter estimation (Audoly et al., 2001). Identifiability
tioning coefficients, membrane permeability coefficients,
problems can also be reduced by decreasing the number of
rate of absorption, etc.). Some parameter values can be
parameters that need to be estimated. Sensitivity analysis
estimated from in vitro experiments (protein-binding rates,
can be used to decide which parameters can be abandoned
Michaelis Menton constants, etc.). Many parameter values
without significantly altering model output. It compares
can be found in the published literature. Some parameters
the relative contributions of reasonable ranges of parame-
can be derived from in vitro and in vivo experimentation.
ter values to an output of interest (Evans and Andersen,
However, there will usually be some parameters for which
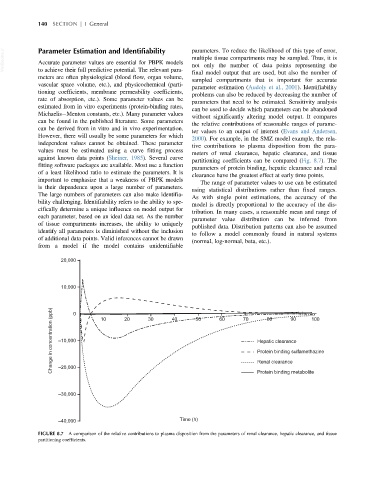
2000). For example, in the SMZ model example, the rela-
independent values cannot be obtained. These parameter
tive contributions to plasma disposition from the para-
values must be estimated using a curve fitting process
meters of renal clearance, hepatic clearance, and tissue
against known data points (Sheiner, 1985). Several curve
partitioning coefficients can be compared (Fig. 8.7). The
fitting software packages are available. Most use a function
parameters of protein binding, hepatic clearance and renal
of a least likelihood ratio to estimate the parameters. It is
clearance have the greatest effect at early time points.
important to emphasize that a weakness of PBPK models
The range of parameter values to use can be estimated
is their dependence upon a large number of parameters.
using statistical distributions rather than fixed ranges.
The large numbers of parameters can also make identifia-
As with single point estimations, the accuracy of the
bility challenging. Identifiability refers to the ability to spe-
model is directly proportional to the accuracy of the dis-
cifically determine a unique influence on model output for
tribution. In many cases, a reasonable mean and range of
each parameter, based on an ideal data set. As the number
parameter value distribution can be inferred from
of tissue compartments increases, the ability to uniquely
published data. Distribution patterns can also be assumed
identify all parameters is diminished without the inclusion
to follow a model commonly found in natural systems
of additional data points. Valid inferences cannot be drawn
(normal, log-normal, beta, etc.).
from a model if the model contains unidentifiable
20,000
10,000 0
Change in concentration (ppb) –10,000 10 20 30 40 50 60 70 Hepatic clearance 100
80
90
Protein binding sulfamethazine
Renal clearance
–20,000
–30,000 Protein binding metabolite
–40,000 Time (h)
FIGURE 8.7 A comparison of the relative contributions to plasma disposition from the parameters of renal clearance, hepatic clearance, and tissue
partitioning coefficients.