Page 168 - Withrow and MacEwen's Small Animal Clinical Oncology, 6th Edition
P. 168
CHAPTER 8 Molecular Diagnostics 147
polymerase chain reaction (PCR)-based methods and high- Tumor DNA Reference DNA
throughput sequencing have allowed us to detect smaller discrete
mutations in DNA that do not involve changes in large portions
VetBooks.ir of the chromosome. Small deletions and insertions in genes, in
addition to single-nucleotide changes, are now routinely detected.
Detection of Chromosomal Abnormalities
Historically these techniques involved the examination of meta-
phase preparations made from chromosomes. The metaphase
preparations were then stained (banded) to help in the identifi-
cation of distinct chromosome morphologies. These techniques
allowed detection of gross abnormalities in the chromosome
number (ploidy) and the presence of chromosomal translocations;
they also led to the identification of genes associated with tumor
development and progression. Cytogenetic analysis has been most
useful in the clinical assessment of leukemias, for which meta-
phase preparations are relatively easy to develop from whole blood
6
samples. For most human leukemias, cytogenetic descriptors are
used to define distinct subgroups into prognostic groups and to
guide treatment decisions.
The use of cytogenetic approaches in the management of com-
panion animals has been limited because of the difficulty involved
in using conventional chromosomal banding to identify canine
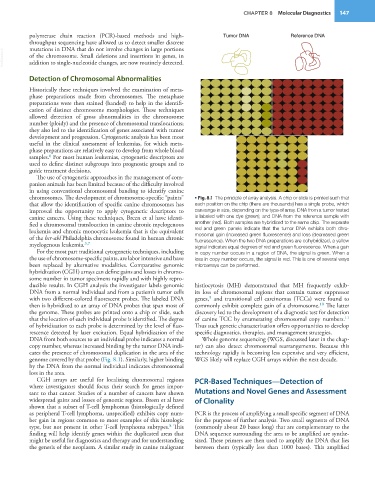
chromosomes. The development of chromosome-specific “paints” • Fig. 8.1 The principle of array analysis. A chip or slide is printed such that
that allow the identification of specific canine chromosomes has each position on the chip (there are thousands) has a single probe, which
improved the opportunity to apply cytogenetic descriptors to can range in size, depending on the type of array. DNA from a tumor tested
canine cancers. Using these techniques, Breen et al have identi- is labeled with one dye (green), and DNA from the reference sample with
fied a chromosomal translocation in canine chronic myelogenous another (red). Both samples are hybridized to the same chip. The separate
leukemia and chronic monocytic leukemia that is the equivalent red and green panels indicate that the tumor DNA exhibits both chro-
of the bcr-abl Philadelphia chromosome found in human chronic mosomal gain (increased green fluorescence) and loss (decreased green
myelogenous leukemia. 3,7 fluorescence). When the two DNA preparations are cohybridized, a yellow
signal indicates equal degrees of red and green fluorescence. When a gain
For the most part traditional cytogenetic techniques, including in copy number occurs in a region of DNA, the signal is green. When a
the use of chromosome-specific paints, are labor intensive and have loss in copy number occurs, the signal is red. This is one of several ways
been replaced by alternative modalities. Comparative genomic microarrays can be performed.
hybridization (CGH) arrays can define gains and losses in chromo-
some number in tumor specimens rapidly and with highly repro-
ducible results. In CGH analysis the investigator labels genomic histiocytosis (MH) demonstrated that MH frequently exhib-
DNA from a normal individual and from a patient’s tumor cells its loss of chromosomal regions that contain tumor suppressor
9
with two different-colored fluorescent probes. The labeled DNA genes, and transitional cell carcinomas (TCCs) were found to
10
then is hybridized to an array of DNA probes that span most of commonly exhibit complete gain of a chromosome. The latter
the genome. These probes are printed onto a chip or slide, such discovery led to the development of a diagnostic test for detection
that the location of each individual probe is identified. The degree of canine TCC by enumerating chromosomal copy numbers.
11
of hybridization to each probe is determined by the level of fluo- Thus such genetic characterization offers opportunities to develop
rescence detected by laser excitation. Equal hybridization of the specific diagnostics, therapies, and management strategies.
DNA from both sources to an individual probe indicates a normal Whole genome sequencing (WGS, discussed later in the chap-
copy number, whereas increased binding by the tumor DNA indi- ter) can also detect chromosomal rearrangements. Because this
cates the presence of chromosomal duplication in the area of the technology rapidly is becoming less expensive and very efficient,
genome covered by that probe (Fig. 8.1). Similarly, higher binding WGS likely will replace CGH arrays within the next decade.
by the DNA from the normal individual indicates chromosomal
loss in the area.
CGH arrays are useful for localizing chromosomal regions PCR-Based Techniques—Detection of
where investigators should focus their search for genes impor-
tant to that cancer. Studies of a number of cancers have shown Mutations and Novel Genes and Assessment
widespread gains and losses of genomic regions. Breen et al have of Clonality
shown that a subset of T-cell lymphomas (histologically defined
as peripheral T-cell lymphoma, unspecified) exhibits copy num- PCR is the process of amplifying a small specific segment of DNA
ber gain in regions common to most examples of this histologic for the purpose of further analysis. Two small segments of DNA
type, but not present in other T-cell lymphoma subtypes. This (commonly about 20 bases long) that are complementary to the
8
finding will help identify genes within the duplicated areas that DNA sequence surrounding the area to be amplified are synthe-
might be useful for diagnostics and therapy and for understanding sized. These primers are then used to amplify the DNA that lies
the genesis of the neoplasm. A similar study in canine malignant between them (typically less than 1000 bases). This amplified