Page 174 - Withrow and MacEwen's Small Animal Clinical Oncology, 6th Edition
P. 174
CHAPTER 8 Molecular Diagnostics 153
With ddPCR, as long as the PCR reaction produces a fluorescent similar to germinal center B cells or similar to activated B cells.
signal, the efficiency with which the reaction achieves that signal Importantly, these two categories had prognostic significance;
germinal center–like DLBCL patients have a better prognosis.
is not as relevant to the results. The technology is also less limited
VetBooks.ir by the amount of sample available. The distinction between germinal center DLBCL and activated
B-cell DLBCL is now well established as a prognostic indicator
Gene Expression Profiling in people. Studies of canine lymphoma have offered conflicting
data with regard to subclassification of canine DLBCL into germi-
Gene expression profiling refers to the quantification of thou- nal center and activated B-cell subtypes based on gene expression
sands of mRNAs simultaneously, to create a picture of global gene profiling, so additional study is necessary. 46,47 Staudt et al made
expression in the cell population. The pattern of gene expression is another important discovery about human B-cell lymphoma,
called a gene expression profile. In most cases gene expression profil- using gene expression profiling, that provided insights into immu-
ing is comparative. Investigators can compare expression profiles nity to cancer. Expression profiles of lymph nodes from patients
of tumor cells to the normal cellular counterpart, to similar tumors with follicular B-cell lymphoma demonstrated that the type of
with different clinical outcomes, to tumor cells from the same immune response signature was prognostic in this disease. Patients
patient before and after therapy, or between primary and meta- whose lymph node gene expression patterns exhibited evidence of
static lesions, to determine how different pathways have changed. T-cell activation had a better overall survival than patients whose
Such information has proven invaluable in a variety of settings. gene expression patterns resembled activated macrophages. The
48
For example, in a study of cervical cancer in people, investiga- expression profile of the tumor cells themselves was not predic-
tors analyzed gene expression profiles of tumors that responded tive. This finding suggests that the immune response contributes
to chemotherapy and radiation and compared those with tumors to survival in patients with follicular B-cell lymphoma, a result
that did not. Their study revealed that activation of the PI3K/Akt corroborated in other types of B-cell lymphoma. 49,50
signaling pathway was associated with a poor response, suggesting
one potential therapeutic target. It is important to note that the Microarrays
44
collective signature of an expression profile can be an informa- Microarrays are solid chips on which DNA probes that are com-
51
tive predictor of disease biology or response to therapy; however, plementary to thousands of different genes are imprinted. Such
alterations in the expression of a single gene are rarely informative platforms expand the study of expression of single genes to a larger
and require extensive post hoc validation. scale needed for an expression signature to be defined. RNA from
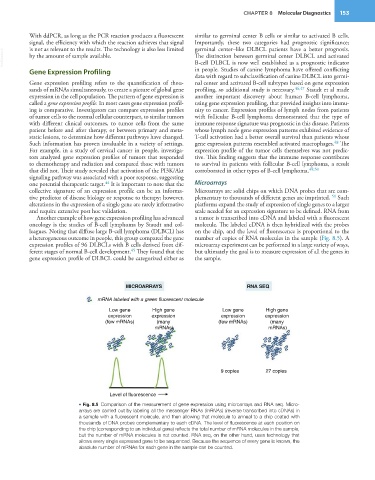
Another example of how gene expression profiling has advanced a tumor is transcribed into cDNA and labeled with a fluorescent
oncology is the studies of B-cell lymphoma by Staudt and col- molecule. The labeled cDNA is then hybridized with the probes
leagues. Noting that diffuse large B-cell lymphoma (DLBCL) has on the chip, and the level of fluorescence is proportional to the
a heterogeneous outcome in people, this group compared the gene number of copies of RNA molecules in the sample (Fig. 8.5). A
expression profiles of 96 DLBCLs with B cells derived from dif- microarray experiment can be performed in a large variety of ways,
45
ferent stages of normal B-cell development. They found that the but ultimately the goal is to measure expression of all the genes in
gene expression profile of DLBCL could be categorized either as the sample.
MICROARRAYS RNA SEQ
mRNA labeled with a green fluorescent molecule
Low gene High gene Low gene High gene
expression expression expression expression
(few mRNAs) (many (few mRNAs) (many
mRNAs) mRNAs)
9 copies 27 copies
Level of fluorescence
• Fig. 8.5 Comparison of the measurement of gene expression using microarrays and RNA seq. Micro-
arrays are carried out by labeling all the messenger RNAs (mRNAs) (reverse transcribed into cDNAs) in
a sample with a fluorescent molecule, and then allowing that molecule to anneal to a chip coated with
thousands of DNA probes complementary to each cDNA. The level of fluorescence at each position on
the chip (corresponding to an individual gene) reflects the total number of mRNA molecules in the sample,
but the number of mRNA molecules is not counted. RNA seq, on the other hand, uses technology that
allows every single expressed gene to be sequenced. Because the sequence of every gene is known, the
absolute number of mRNAs for each gene in the sample can be counted.