Page 120 - Science
P. 120
RESEARCH | REPORT
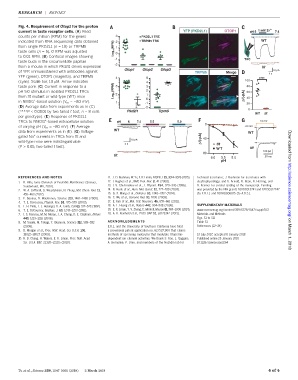
Fig. 4. Requirement of Otop1 for the proton
current in taste receptor cells. (A) Read YFP (PKD2L1) OTOP1 pH 5 1mMZn 2+ 7.4
counts per million (RPM) for the genes 3 PKD2L1 TRC
indicated from RNA-sequencing data obtained 2 TRPM5 TRC WT
from single PKD2L1 (n = 19) or TRPM5 (RPM) 1
taste cells (n = 5). 0 RPM was adjusted Log 10 0
to 0.01 RPM. (B) Confocal images showing
-1 TRC
taste buds in the circumvallate papillae
-2
from a mouse in which Pkd2l1 drives expression tlt
Otop1 Otop2 Otop3
of YFP, immunostained with antibodies against TRPM5 Merge 10 pA
2s
YFP (green), OTOP1 (magenta), and TRPM5 4 5 ****
(cyan). Scale bar, 10 mM. Arrow indicates 4 100
taste pore. (C) Current in response to a (RPM) 3 3 80
pH 5.0 stimulus in isolated PKD2L1 TRCs 2 2 60
from tlt mutant or wild-type (WT) mice Log 10 1 0 ΔI(-pA) 40
+
in NMDG -based solution (V m = –80 mV). 1 -1 20
(D) Average data from experiments as in (C) -2
(****P< 0.0001 by two tailed t test, n = 8 cells Pkd2l1 Trpm5 0
WT tlt
per genotype). (E) Response of PKD2L1
+
TRCs to NMDG -based extracellular solution pH 6 7.4 5.5 5.0 10 mV
0
of varying pH (V m = –80 mV). (F)Average -80 mV
WT
data from experiments as in (E). (G)Voltage- WT
+
gated Na currents in TRCs from tlt and
10 pA ΔI(-pA) 40
wild-type mice were indistinguishable
5s tlt
(P > 0.05, two-tailed t test). tlt Downloaded from
WT
100 pA
80
tlt
6.0 5.5 5.0 20 ms
pH
REFERENCES AND NOTES 11. J. D. Bushman, W. Ye, E. R. Liman, FASEB J. 29,3014–3026 (2015). technical assistance; J. Bushman for assistance with
1. B. Hille, Ionic Channels of Excitable Membranes (Sinauer, 12. I. Hughes et al., BMC Evol. Biol. 8, 41 (2008). electrophysiology; and D. Arnold, B. Bean, B. Herring, and
Sunderland, MA, 2001). 13. I. V. Chizhmakov et al., J. Physiol. 494,329–336 (1996). R. Kramer for careful reading of the manuscript. Funding
2. W. A. Catterall, G. Wisedchaisri, N. Zheng, Nat. Chem. Biol. 13, 14. B. Hurle et al., Hum. Mol. Genet. 12, 777–789 (2003). was provided by the NIH grants R01DC013741 and R21DC012747 http://science.sciencemag.org/
455–463 (2017). 15. G. X. Wang et al., Diabetes 63, 1340–1352 (2014). (to E.R.L.) and R01HG006015 (to A.D.S.).
3. E. Gouaux, R. Mackinnon, Science 310,1461–1465 (2005). 16. C. Wu et al., Genome Biol. 10, R130 (2009).
4. T. E. Decoursey, Physiol. Rev. 83, 475–579 (2003). 17. E. Kim et al., Mol. Cell. Neurosci. 46, 655–661 (2011).
5. L. H. Pinto, L. J. Holsinger, R. A. Lamb, Cell 69,517–528 (1992). 18. A. L. Huang et al., Nature 442, 934–938 (2006). SUPPLEMENTARY MATERIALS
6. T. E. DeCoursey, Biophys. J. 60, 1243–1253 (1991). 19. E. R. Liman, Y. V. Zhang, C. Montell, Neuron 81,984–1000 (2014). www.sciencemag.org/content/359/6379/1047/suppl/DC1
7. I. S. Ramsey, M. M. Moran, J. A. Chong, D. E. Clapham, Nature 20. A. K. Reinhold et al., PLOS ONE 10, e0123342 (2015). Materials and Methods
440, 1213–1216 (2006). Figs. S1 to S11
8. M. Sasaki, M. Takagi, Y. Okamura, Science 312, 589–592 ACKNOWLEDGMENTS Table S1
(2006). E.R.L. and the University of Southern California have filed References (21–34)
9. D. Morgan et al., Proc. Natl. Acad. Sci. U.S.A. 106, a provisional patent application no. 62/537,900 that claims
18022–18027 (2009). methods of screening molecules that modulate Otopetrin- 13 July 2017; accepted 8 January 2018 on March 1, 2018
10. R. B. Chang, H. Waters, E. R. Liman, Proc. Natl. Acad. dependent ion channel activities. We thank S. Rao, L. Goggins, Published online 25 January 2018
Sci. U.S.A. 107, 22320–22325 (2010). A. Bernanke, P. Uren, and members of the Nuzhdin lab for 10.1126/science.aao3264
Tu et al., Science 359, 1047–1050 (2018) 2 March 2018 4of4